单细胞分析一直是生信以及医学期刊比较流行的方向,小果在浏览期刊的时候发现了这样一个R包—ScPP(Single Cells’Phenotype Prediction),这个R包是一种基于表型相关标记基因在整体和单细胞中的表达谱,来识别具有特定表型的细胞亚群的工具,是一个于2023年新发布的单细胞表型预测工具,可以非常快速的进行单细胞的相关分析。话不多说,小果来带大家解锁ScPP这个包的用法!(本次所用R包对服务器性能要求较高,欢迎小伙伴们来租赁我们的服务器来运行代码呦~)
在进行教程之前,我们首先需要了解什么是单细胞表型预测,单细胞表型预测(Single Cells’Phenotype Prediction)是一种利用单细胞数据来推断细胞的功能或状态的方法,可以帮助研究人员发现细胞的异质性、转化关系,并且有助于揭示细胞表型与基因型、蛋白质以及环境因素之间的关联,还可以应用于药物响应、疾病诊断、组织发育和干细胞分化等。可以说是单细胞分析中非常具有开发潜能的一个方向呢。
我们在进行单细胞表型预测时通常需要以下几个步骤:
1.从单细胞数据中提取特征,比如说基因表达、表观遗传修饰、蛋白质水平或细胞形态等特征。
2.选择与表型相关的特征,可以通过差异分析、特征选择或深度学习等方法进行筛选。
3.建立预测模型,这一步可以通过监督学习、无监督学习或半监督学习等方法完成。
4.验证和评估预测结果,通过交叉验证、ROC曲线、混淆矩阵或生物学实验等方法进行验证。
了解了什么是单细胞表型预测,我们来看一下今天的主角——ScPP。下面的图片是通过ScPP进行单细胞表型预测的流程图:
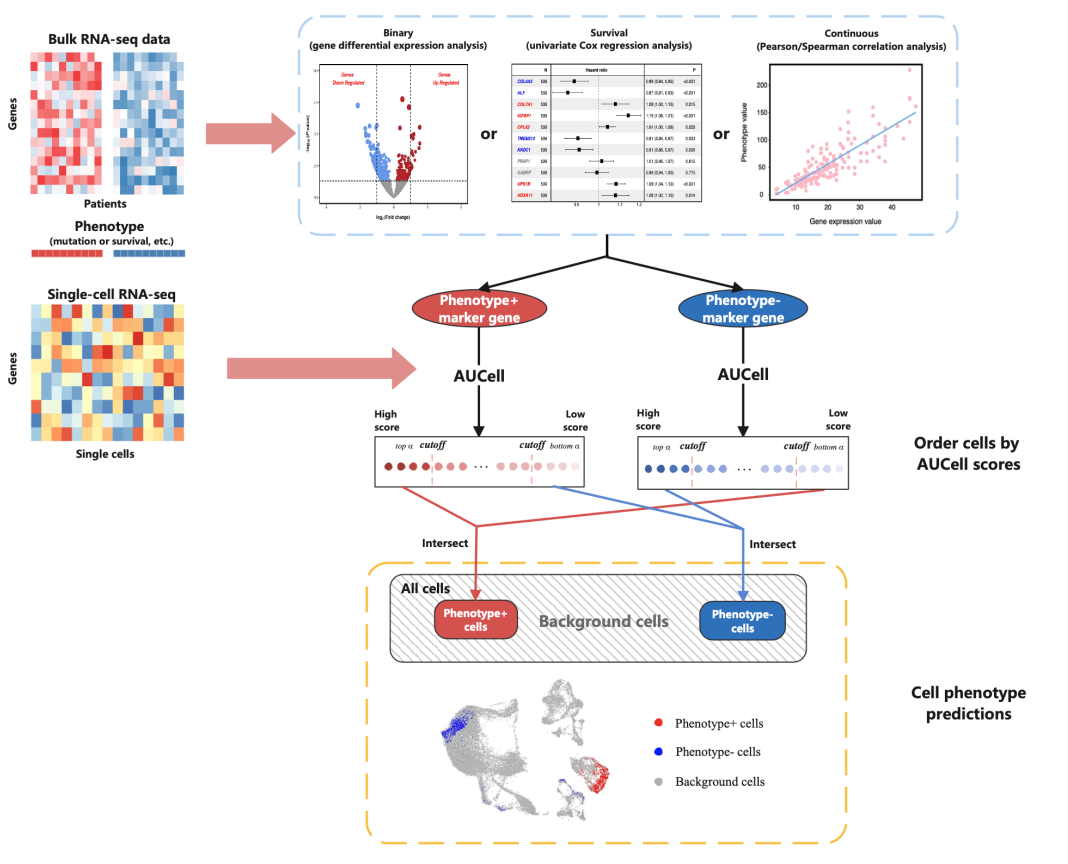
首先,我们可以通过Bulk RNA-seq获取一组细胞的转录组测序数据,然后运用Binary进行差异表达分析,当然也可以使用生存分析或者连续变量相关性分析,提取出Phenotype+ 标记基因以及Phenotype-标记基因,进而将其用于鉴定和分类细胞。
除了Bulk RNA-seq,也可以直接进行Single-cell RNA-seq,找到Phenotype标记基因,然后进行AUCell分析,识别具有标记基因的细胞群,并且将Phenotype-以及Phenotype+区分的结果进行合并,就可以得到在整个细胞中,表型基因表达高和表达低的分布图,也就完成了表型预测。
接下来是代码实操:
Step 1 进行ScPP包的初始化
#导入ScPP中的功能函数
##为单细胞表达数据进行预处理。输入数据是一个计数矩阵,其中细胞是列名,基因是行名
sc_Preprocess <- function(counts, project = “sc_preprocess”,
normalization.method = “LogNormalize”, scale.factor = 10000,
selection.method = “vst”, resolution = 0.1,
dims_Neighbors = 1:20, dims_TSNE = 1:20, dims_UMAP = 1:20){
library(Seurat)
sc_dat <- CreateSeuratObject(counts = counts, project = project)
sc_dat <- NormalizeData(object = sc_dat, normalization.method = normalization.method, scale.factor = scale.factor)
sc_dat <- FindVariableFeatures(object = sc_dat, selection.method = selection.method)
sc_dat <- ScaleData(object = sc_dat, features = rownames(sc_dat))
sc_dat <- RunPCA(object = sc_dat, features = VariableFeatures(sc_dat))
sc_dat <- FindNeighbors(object = sc_dat, dims = dims_Neighbors)
sc_dat <- FindClusters(object = sc_dat, resolution = resolution)
sc_dat <- RunTSNE(object = sc_dat, dims = dims_TSNE)
sc_dat <- RunUMAP(object = sc_dat, dims = dims_UMAP)
return(sc_dat)
}
#运用Binary进行差异表达分析
##生成二元变量与表型+或表型-组相关的标记基因以及特征,一共包含4个参数:bulk_data、features、ref_group和Log2FC_cutoff
marker_Binary <- function(bulk_data, features, ref_group, Log2FC_cutoff = 0.585){
library(dplyr)
if (missing(ref_group))
stop(“‘ref_group’ is missing or incorrect.”)
if (missing(bulk_data) || !class(bulk_data) %in% c(“matrix”, “data.frame”))
stop(“‘bulk_data’ is missing or incorrect.”)
if (missing(features) || !class(features) %in% c(“matrix”, “data.frame”))
stop(“‘features’ is missing or incorrect.”)
ref = features$Sample[features$Feature == ref_group]
tes = features$Sample[features$Feature != ref_group]
ref_pos = which(colnames(bulk_data) %in% ref)
tes_pos = which(colnames(bulk_data) %in% tes)
pvalues <- apply(bulk_data, 1, function(x) {
t.test(as.numeric(x)[tes_pos], as.numeric(x)[ref_pos])$p.value
})
log2FCs <- rowMeans(bulk_data[, tes_pos]) – rowMeans(bulk_data[, ref_pos])
res <- data.frame(pvalue = pvalues, log2FC = log2FCs)
res <- res[order(res$pvalue), ]
res$fdr <- p.adjust(res$pvalue, method = “fdr”)
geneList <- list(
gene_pos = res %>% filter(pvalue < 0.05, log2FC > Log2FC_cutoff ) %>% rownames(.),
gene_neg = res %>% filter(pvalue < 0.05, log2FC < -Log2FC_cutoff ) %>% rownames(.)
)
if(length(geneList[[1]]) > 0 & length(geneList[[2]] > 0)){return(geneList)}
else if (length(geneList[[1]]) == 0){warning(“There is no genes positively correlated with the given feature in this bulk dataset.”);geneList = list(gene_pos = geneList[[2]]);return(geneList)}
else if (length(geneList[[2]]) == 0){warning(“There is no genes negatively correlated with the given feature in this bulk dataset.”);geneList = list(gene_neg = geneList[[1]]);return(geneList)}
}
#进行连续变量相关性分析
##生成连续变量以及和表型相关的标记基因或特征,包含4个参数:bulk_data、features、method和estimate_cutoff
marker_Continuous <- function(bulk_data, features, method = “spearman”, estimate_cutoff = 0.2){
library(dplyr)
if (missing(bulk_data) || !class(bulk_data) %in% c(“matrix”, “data.frame”))
stop(“‘bulk_data’ is missing or incorrect.”)
if (missing(features) || !class(features) %in% c(“integer”, “numeric”))
stop(“‘features’ is missing or incorrect.”)
CorrelationTest = apply(bulk_data,1,function(x){
pvalue = cor.test(as.numeric(x),log2(as.numeric(features)+1), method = method)$p.value
estimate = cor.test(as.numeric(x),log2(as.numeric(features)+1), method = method)$estimate
res = cbind(pvalue,estimate)
return(res)
})
resc = t(CorrelationTest)
colnames(resc) = c(“pvalue”,”estimate”)
resc = as.data.frame(resc[order(as.numeric(resc[,1]),decreasing = FALSE),])
resc$fdr <- p.adjust(resc$pvalue,method = “fdr”)
geneList <- list(
gene_pos = resc %>% filter(fdr < 0.05, estimate > estimate_cutoff) %>% rownames(.),
gene_neg = resc %>% filter(fdr < 0.05, estimate < -estimate_cutoff) %>% rownames(.)
)
if(length(geneList[[1]]) > 0 & length(geneList[[2]] > 0)){return(geneList)}
else if (length(geneList[[1]]) == 0){warning(“There is no genes positively correlated with the given feature in this bulk dataset.”);geneList = list(gene_pos = geneList[[2]]);return(geneList)}
else if (length(geneList[[2]]) == 0){warning(“There is no genes negatively correlated with the given feature in this bulk dataset.”);geneList = list(gene_neg = geneList[[1]]);return(geneList)}
}
##生存数据分析,生成与患者预后相关的标记基因或特征,包含2个参数:bulk_data和survival_data
marker_Survival <- function(bulk_data,survival_data){
library(survival)
library(dplyr)
if (missing(bulk_data) || !class(bulk_data) %in% c(“matrix”, “data.frame”))
stop(“‘bulk_data’ is missing or incorrect.”)
if (missing(survival_data) || !class(survival_data) %in% c(“matrix”, “data.frame”))
stop(“‘survival_data’ is missing or incorrect.”)
SurvivalData <- data.frame(cbind(survival_data,t(bulk_data)))
colnames(SurvivalData) = make.names(colnames(SurvivalData))
var <- make.names(rownames(bulk_data))
Model_Formula <- sapply(var, function(x) as.formula(paste(“Surv(time, status) ~”, x)))
Model_all <- lapply(Model_Formula, function(x) coxph(x, data = SurvivalData))
res <- lapply(seq_along(Model_all), function(i) {
coef_summary <- summary(Model_all[[i]])$coefficients
data.frame(
variable = var[i],
pvalue = coef_summary[,5],
coef = coef_summary[,2]
)
}) %>% bind_rows()
res <- res[order(res$pvalue), ]
res$fdr <- p.adjust(res$pvalue, method = “fdr”)
geneList <- list(
gene_pos = res %>% filter(fdr < 0.05, coef > 1) %>% pull(variable), #correalted with worse survival
gene_neg = res %>% filter(fdr < 0.05, coef < 1) %>% pull(variable) #correlated with better survival
)
if(length(geneList[[1]]) > 0 & length(geneList[[2]] > 0)){return(geneList)}
else if (length(geneList[[1]]) == 0){warning(“There is no genes negatively correlated with patients’ prognosis in this bulk dataset.”);geneList = list(gene_pos = geneList[[2]]);return(geneList)}
else if (length(geneList[[2]]) == 0){warning(“There is no genes positively correlated with patients’ prognosis in this bulk dataset.”);geneList = list(gene_neg = geneList[[1]]);return(geneList)}
}
##进行单细胞表型预测,包含3个参数:sc_dataset, geneList和probs。
ScPP = function(sc_dataset, geneList, probs = 0.2){
if(length(geneList) != 2){
stop(“This gene list do not have enough information correlated with interested feature.”)
}
if (missing(sc_dataset) || class(sc_dataset) != “Seurat”)
stop(“‘sc_dataset’ is missing or not a seurat object.”)
library(AUCell)
cellrankings = AUCell_buildRankings(sc_dataset@assays$RNA@data,plotStats = FALSE)
cellAUC = AUCell_calcAUC(geneList,cellrankings)
metadata = as.data.frame(sc_dataset@meta.data)
metadata$AUCup <- as.numeric(getAUC(cellAUC)[“gene_pos”, ])
metadata$AUCdown <- as.numeric(getAUC(cellAUC)[“gene_neg”, ])
downcells1 = rownames(metadata)[which(metadata$AUCup <= quantile(metadata$AUCup,probs = probs))]
upcells1 = rownames(metadata)[which(metadata$AUCup >= quantile(metadata$AUCup,probs = (1-probs)))]
downcells2 = rownames(metadata)[which(metadata$AUCdown >= quantile(metadata$AUCdown,probs = (1-probs)))]
upcells2 = rownames(metadata)[which(metadata$AUCdown <= quantile(metadata$AUCdown,probs = probs))]
ScPP_neg = intersect(downcells1,downcells2)
ScPP_pos = intersect(upcells1,upcells2)
metadata$ScPP <- ifelse(rownames(metadata) %in% ScPP_pos, “Phenotype+”, “Background”)
metadata$ScPP <- ifelse(rownames(metadata) %in% ScPP_neg, “Phenotype-“, metadata$ScPP)
return(metadata)
}
#导入绘图所需的包
library(ggplot2)
#安装Scpp包以及相关的R包
if (!require(“BiocManager”, quietly = TRUE))
install.packages(“BiocManager”)
BiocManager::install(“AUCell”)
if (!requireNamespace(“devtools”, quietly = TRUE))
install.packages(“devtools”)
devtools::install_github(“WangX-Lab/ScPP”)
Step 2 应用二元变量进行ScPP
本次分析所需的数据小果已经帮大家整理好了,大家点击链接即可下载:
链接:https://pan.baidu.com/s/1Kuox0OuispGobwKDeQJ0Bw
提取码:v4fz
# 准备数据
load(“/data/binary.RData”)
# 数据总览
bulk[1:6,1:6] # bulk data
head(binary) # phenotype data of bulk
sc_count[1:6,1:6] # scRNA-seq data
# 通过 ScPP 选择信息细胞
sc = sc_Preprocess(sc_count)
geneList = marker_Binary(bulk, binary, ref_group = “Normal”)
metadata = ScPP(sc, geneList)
write.csv(metadata, “/results/binary_meta.csv”)
head(metadata)
sc$ScPP = metadata$ScPP
Idents(sc) = “ScPP”
#对ScPP识别的细胞进行可视化
DimPlot(sc, group = “ScPP”, cols = c(“grey”,”blue”,”red”))
ggsave(“Fig_binary.pdf”,path = “/results/”,width = 10, height = 10, units = “cm”)
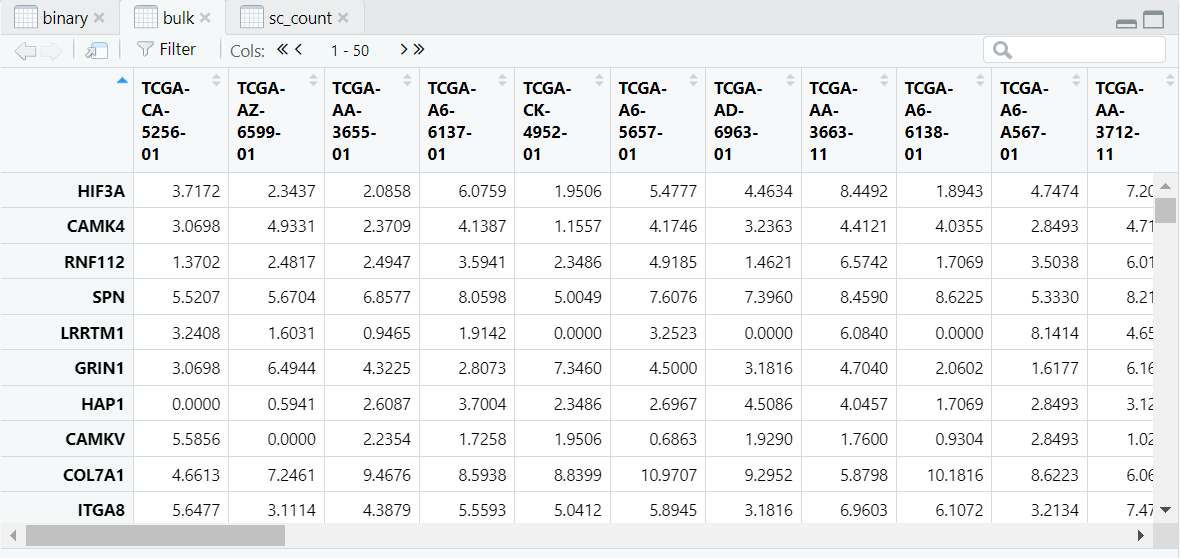
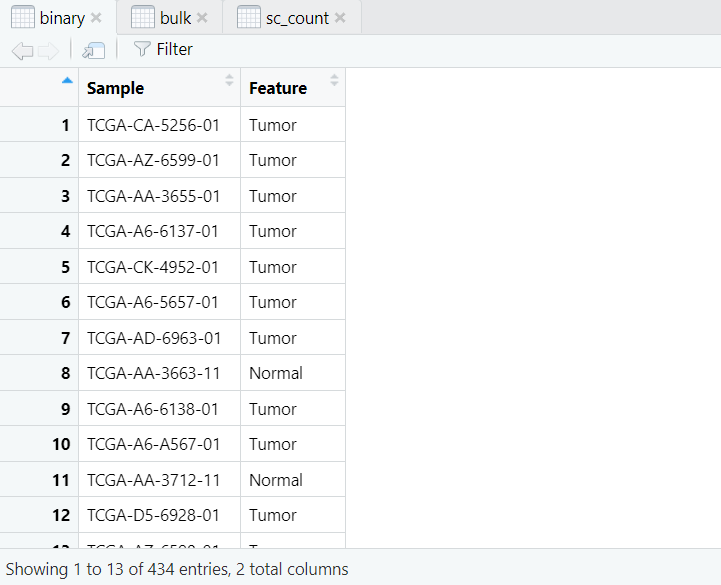
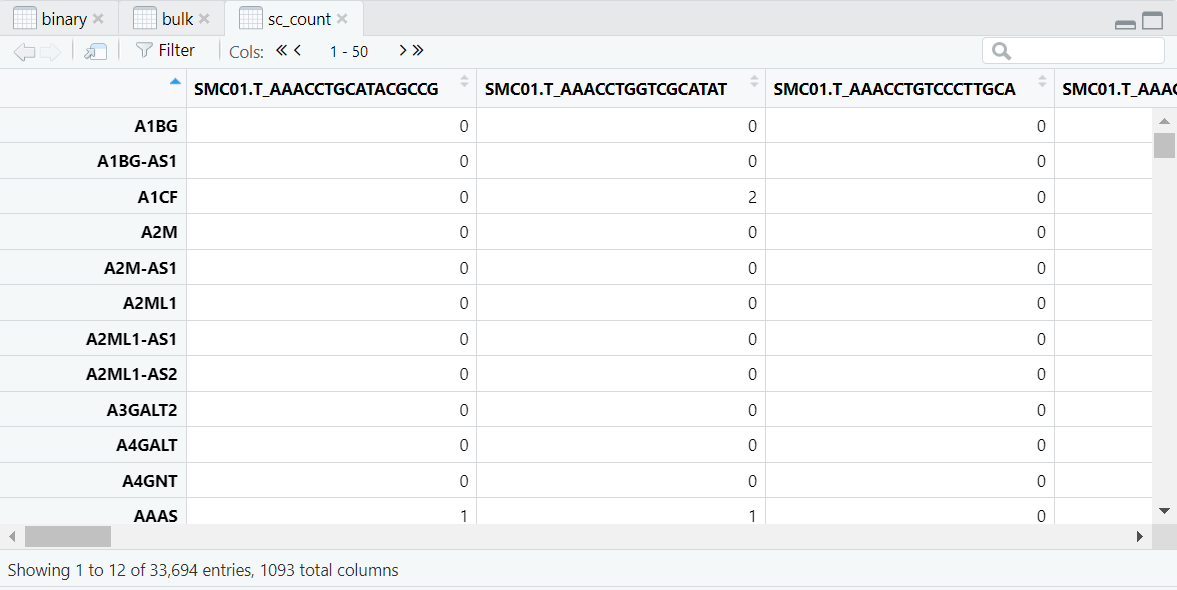
导入的数据如图,是我们进行Binary分析所需的三个数据,分别是:Binary数据(样本的肿瘤与正常的二元数据),Bulk数据(表型数据),sc_count数据(scRNA的测序数据)。
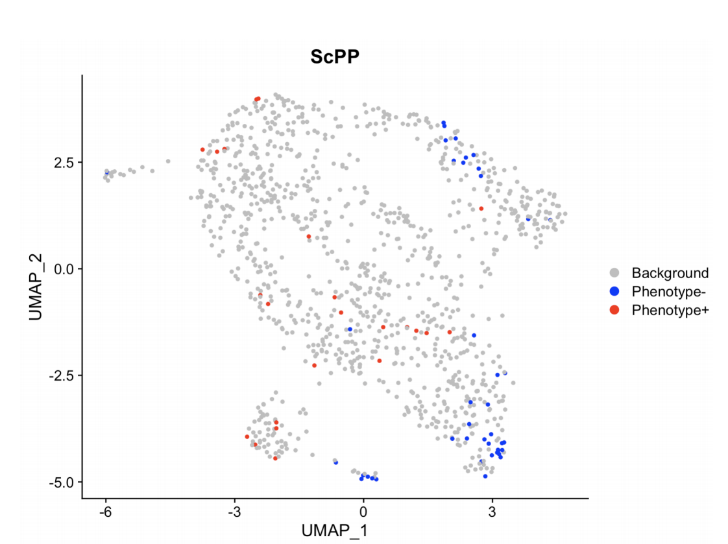
接着是我们分析的结果,通过ScPP利用二元变量分析得到了细胞中的背景基因,以及带有标记基因的高表达和低表达群,图中的红色表示高表达群,说明这部分的细胞可能是具有我们所需要的表型的细胞。
Step 3 运用连续变量进行ScPP
# 导入数据(所需数据小果放在上文的链接了哦~)
# library(ScPP)
load(“/data/continuous.RData”)
# 数据总览
bulk[1:6,1:6] # bulk data
head(continuous) # phenotype data of bulk
sc_count[1:6,1:6] # scRNA-seq data
# 通过 ScPP 选择信息细胞
sc = sc_Preprocess(sc_count)
geneList = marker_Continuous(bulk, continuous$TMB_non_silent)
metadata = ScPP(sc, geneList)
write.csv(metadata, “/results/continuous_meta.csv”)
sc$ScPP = metadata$ScPP
Idents(sc) = “ScPP”
#对ScPP识别的细胞进行可视化
DimPlot(sc, group = “ScPP”, cols = c(“grey”,”blue”,”red”))
ggsave(“Fig_continuous.pdf”,path = “/results/”,width = 10, height = 10, units = “cm”)
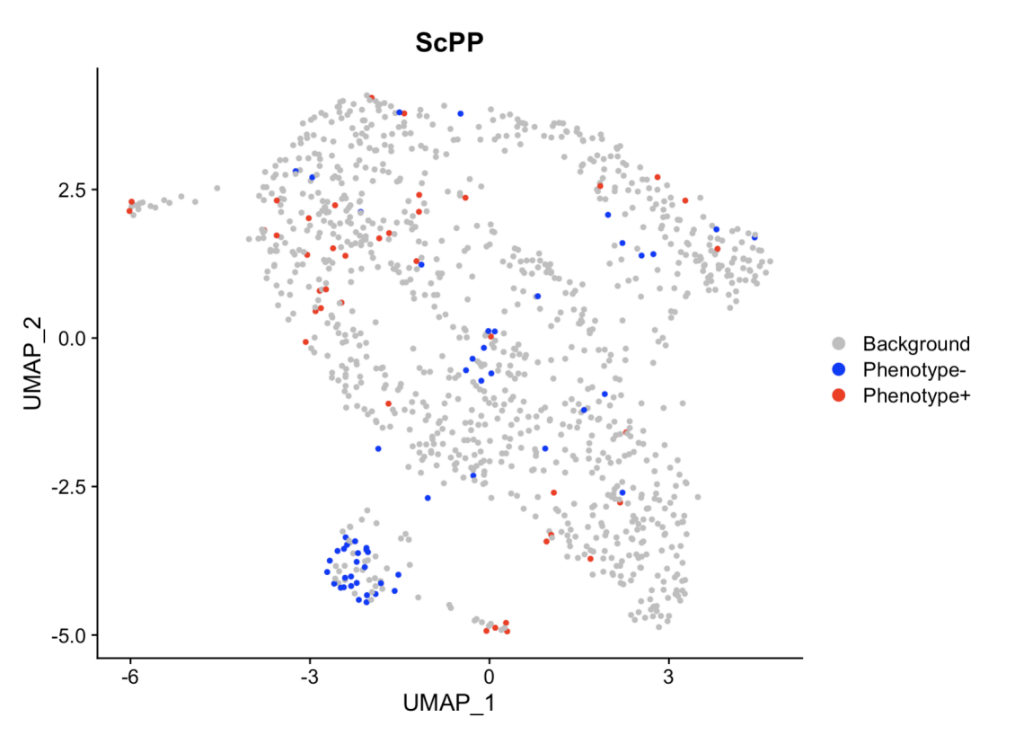
结果的可视化如图,可以发现和运用二元变量进行的ScPP的结果是比较相似的,这也可以证明ScPP这个包的分析比较稳定。ScPP的分析文件储存在csv文件中,小伙伴们也可以用csv文件进行其他的分析~
Step 4 运用生存数据进行ScPP
#导入数据
load(“/data/survival.RData”)
#通过 ScPP 选择信息细胞
sc = sc_Preprocess(sc_count)
geneList = marker_Survival(bulk, survival)
str(geneList)
metadata = ScPP(sc, geneList)
write.csv(metadata, “/results/survival_meta.csv”)
sc$ScPP = metadata$ScPP
Idents(sc) = “ScPP”
#可视化
DimPlot(sc, group = “ScPP”, cols = c(“grey”,”blue”,”red”))
ggsave(“Fig_survival.pdf”,path = “/results/”,width = 10, height = 10, units = “cm”)
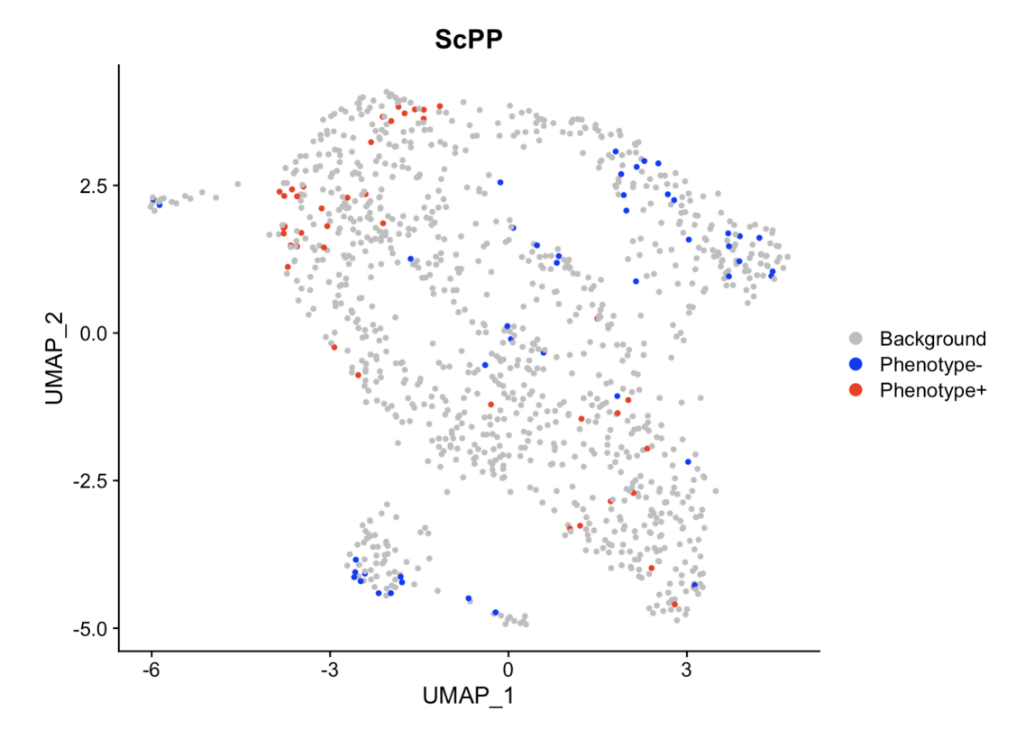
结果如图,可以发现我们进行ScPP的结果与前两次的分析略有不同,但是整体的细胞表型分布还是大致相同的。
以上就是小果今天分享的全部内容啦!今天我们主要了解了什么是单细胞表型预测,以及运用三种数据(二进制数据,生存数据,连续性数据)在ScPP这个R包的帮助下进行单细胞表型预测,并且对结果进行了解读。如果大家对今天的分享有任何疑问,欢迎关注公众号向小果提问,我们有专门的生信分析服务哦。同时,如果大家没有做出来图也不要灰心,可以试一试我们的云生信小工具哦,只要输入合适的数据以及指令就可以直接绘制想要的图呢,链接:http://www.biocloudservice.com/home.html。