今天小果利用ggcor包绘制好看的相关性图,代码如下:
- 安装需要的R包
install.packages(“ggplot2”)
install.packages(“GSVA”)
install.packages(“data.table”)
install.packages(“ggnewscale”)
install.packafes(“ade4”)
#直接下载安装包安装
install.packages(“ggcor-master.tar.gz”, repos = NULL, type = “source”)
- 导入需要的R包
library(ggplot2)
library(ggcor)
library(ggplot2)
library(data.table)
library(GSVA)
- 代码展示
#导入需要的数据
#加载TCGA-BLCA表达谱FPKM
expr<-read.table(“tcga_blca_fpkm.txt”,sep = “\t”,row.names = 1,check.names = F,stringsAsFactors = F,header = T)
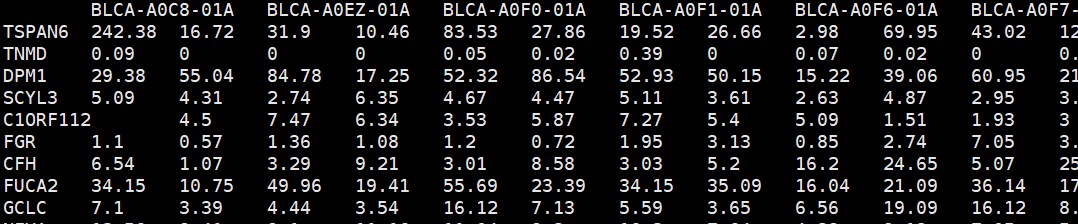
tumsam <- colnames(expr)[substr(colnames(expr),11,13) == “01A”] # 取出肿瘤样本
# 取出SIGLEC15表达量
siglec15 <- log2(expr[“SIGLEC15”,tumsam] + 1)
write.csv(siglec15, “gene.csv”)
# 加载immunotherapy-predicted pathways
immPath <- read.table(“immunotherapy predicted pathways.txt”,sep = “\t”,row.names = 1,check.names = F,stringsAsFactors = F,header = T)
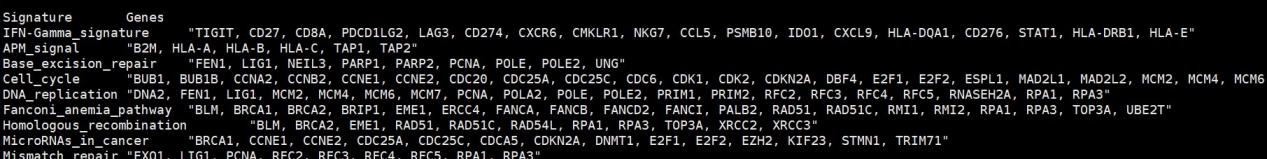
immPath.list <- list()
for (i in rownames(immPath)) {
tmp <- immPath[i,”Genes”]
tmp <- toupper(unlist(strsplit(tmp,”, “,fixed = T)))
tmp <- gsub(” “,””,tmp)
immPath.list[[i]] <- tmp
}
# 计算immunotherapy-predicted pathways的富集得分
immPath.score <- gsva(expr = as.matrix(log2(expr[,tumsam] + 1)),
immPath.list,
method = “ssgsea”)
write.table(immPath.score, “immPath.txt”,sep = “\t”,row.names = T,col.names = NA,quote = F)
# 加载单个基因的表达量,也就是“1对多”的“1”
siglec15 <- read.csv(“gene.csv”, row.names = 1, check.names = F)
# 加载富集得分
immPath.score <- read.table(“immPath.txt”, check.names = F)
# 跟目标基因SIGLEC15的表达量合并
immPath.score <- rbind.data.frame(immPath.score,
siglec15)
# 循环计算相关性并绘制左下角
immCorSiglec15 <- NULL
for (i in rownames(immPath.score)) {
cr <- cor.test(as.numeric(immPath.score[i,]),
as.numeric(siglec15),
method = “pearson”)
immCorSiglec15 <- rbind.data.frame(immCorSiglec15,
data.frame(gene = “Siglec15″,
path = i,
r = cr$estimate,
p = cr$p.value,
stringsAsFactors = F),
stringsAsFactors = F)
}
immCorSiglec15$sign <- ifelse(immCorSiglec15$r > 0,”pos”,”neg”)
immCorSiglec15$absR <- abs(immCorSiglec15$r)
immCorSiglec15$rSeg <- as.character(cut(immCorSiglec15$absR,c(0,0.25,0.5,0.75,1),labels = c(“0.25″,”0.50″,”0.75″,”1.00”),include.lowest = T))
immCorSiglec15$pSeg <- as.character(cut(immCorSiglec15$p,c(0,0.001,0.01,0.05,1),labels = c(“<0.001″,”<0.01″,”<0.05″,”ns”),include.lowest = T))
immCorSiglec15[nrow(immCorSiglec15),”pSeg”] <- “Not Applicable”
immCorSiglec15$rSeg <- factor(immCorSiglec15$rSeg, levels = c(“0.25″,”0.50″,”0.75″,”1.00”))
immCorSiglec15$pSeg <- factor(immCorSiglec15$pSeg, levels = c(“<0.001″,”<0.01″,”<0.05″,”Not Applicable”,”ns”))
immCorSiglec15$sign <- factor(immCorSiglec15$sign, levels = c(“pos”,”neg”))
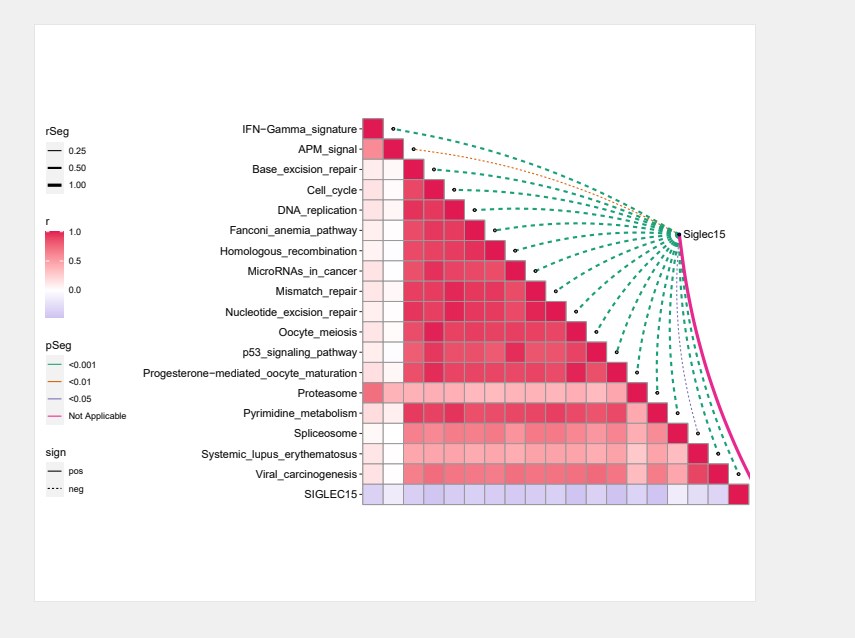
p1 <- quickcor(t(immPath.score),
type = “lower”,
show.diag = TRUE) +
geom_colour() +
add_link(df = immCorSiglec15,
mapping = aes(colour = pSeg, size = rSeg, linetype = sign),
spec.key = “gene”,
env.key = “path”,
diag.label = FALSE) +
scale_size_manual(values = c(0.5, 1, 1.5, 2)) +
scale_color_manual(values = c(“#19A078″,”#DA6003″,”#7570B4″,”#E8288E”,”#65A818″)) +
scale_fill_gradient2(low = “#9483E1”,mid = “white”,high = “#E11953”,midpoint=0) +
remove_axis(“x”)
ggsave(filename = “ggcor.pdf”, width = 10,height = 8)
#immPath.txt-富集得分文件
最终小果成功的画出了相关性网络图,看起来小果非常不错,有需要的可以借鉴学习奥。