今天小果分享一下免疫浸润ssGESA分析,从数据下载到分析,代码如下:
- 安装需要的R包
install.packages(“ggplot2”)
Install.packages(“tidyverse”)
install.packages(“GSVA”)
install.packages(“pheatmap”)
- 载入需要的R包
library(ggplot2)
library(tidyverse)
library(GSVA)
library(pheatmap)
- 数据下载
在Xena数据库下载表达矩阵和ID对应表格
#表达矩阵下载
wget https://gdc-hub.s3.us-east-1.amazonaws.com/download/TCGA-KIRC.htseq_fpkm.tsv.gz
#基因ID转化列表
wget https://gdc-hub.s3.us-east-1.amazonaws.com/download/gencode.v22.annotation.gene.probeMap
- 代码展示
#基因ID转换
expr<-read.table(“TCGA-KIRC.tsv”,header=T,sep=”\t”,row.names=1)
geneann<-read.table(“gencode.v22.annotation.gene.probeMap”,header=T,sep=”\t”,row.names=1)
expr$gsym <- geneann[rownames(expr),]$gene
expr<-distinct(expr,gsym,.keep_all=T)
row.names(expr)<-expr$gsym
expr<-slect(expr,-gsym)
write.table(expr,”TCGA-KIRC-expr.txt”,col.names=T,row.names=T,sep=”\t”)
#ssGSEA分析
exp<-read.table(“TCGA-KIRC-expr.txt”,header=T,row.names=1,sep=”\t”)
geneset<-read.table(“genelist.txt”,header=T,sep=”\t”)
构建免疫细胞Marker基因集:
geneset2 <- split(geneset$Metagene,geneset$Cell.type)
#ssGSEA:
res <- gsva(as.matrix(exp),
geneset2,
method = “ssgsea”,
kcdf = “Gaussian”,
mx.diff = F,
verbose = F)
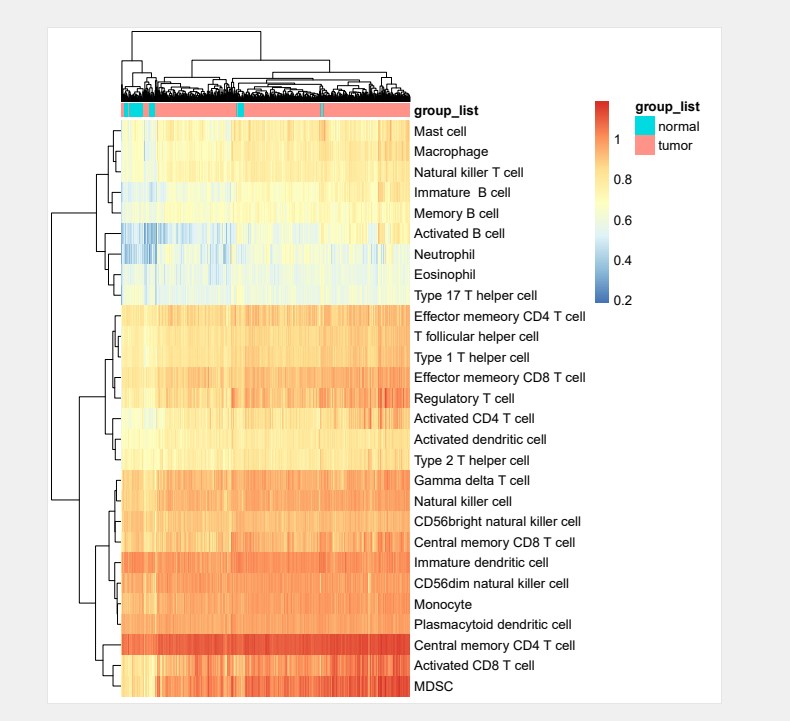
#给热图添加样本注释信息:
group_list <- ifelse(str_sub(colnames(res),14,15) < 10,”tumor”,”normal”)
annotation <- data.frame(group_list)
rownames(annotation) <- colnames(res)
pheatmap(res,
show_colnames = F,
annotation_col = annotation,
fontsize = 10,
filename=”ssGSEA.pdf”)
今天小果的分享就到这里,有需要的可以借鉴学习。