三分钟学会如何绘制聚类+基因表达趋势+功能注释组合图
今天小果想安利一个R包ClusterGVis,分享一下如何利用ClusterGVis包绘制聚类+基因表达趋势+功能注释组合图,下面进行操作,代码如下:
- 安装需要的R包
install.packages(“devtools”)
devtools::install_github(“junjunlab/ClusterGVis”)
install.packages(“Biobase”)
- 载入需要的R包
library(ClusterGVis)
library(Biobase)
- 代码展示
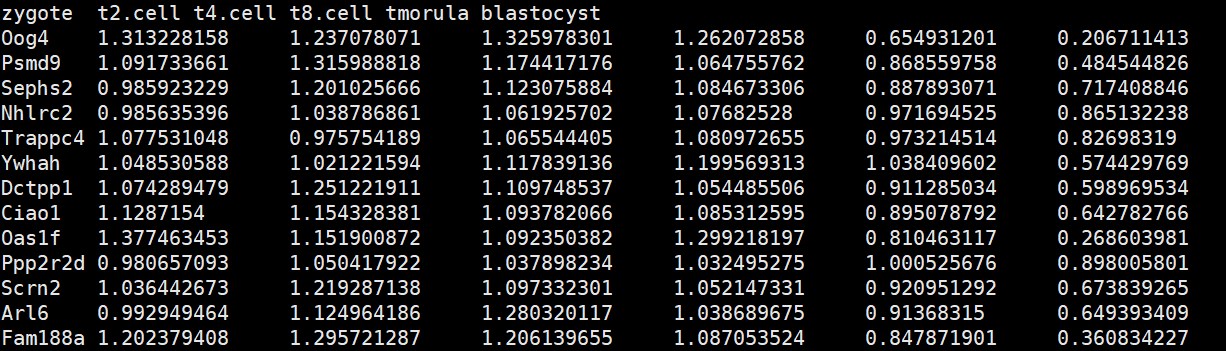
#数据读取-基因表达矩阵文件
expr <- read.table(“exp.txt”,header = T,row.names = 1,sep = “\t”)
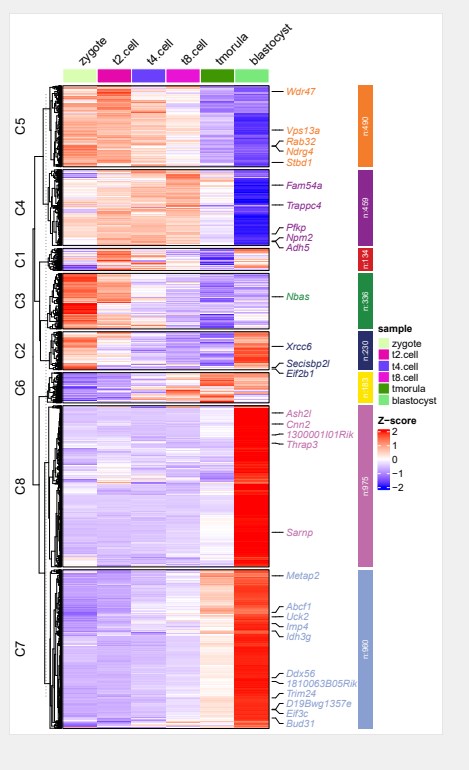
#开始绘图
ck <- clusterData(exp = expr,
cluster.method = “kmeans”,
cluster.num = 8)
#需要标记的基因名称
markGenes = rownames(expr)[sample(1:nrow(expr),30,replace = F)]
pdf(‘clustergene.pdf’,height = 10,width = 6,onefile = F)
visCluster(object = ck,
plot.type = “heatmap”,
column_names_rot = 45,
markGenes = markGenes)
dev.off()
library(org.Mm.eg.db)
# enrich for clusters
enrich <- enrichCluster(object = ck,
OrgDb = org.Mm.eg.db,
type = “BP”,
pvalueCutoff = 0.05,
topn = 5,
seed = 5201314)
pdf(‘enrich.pdf’,height = 10,width = 11,onefile = F)
visCluster(object = ck,
plot.type = “both”,
column_names_rot = 45,
show_row_dend = F,
markGenes = markGenes,
markGenes.side = “left”,
genes.gp = c(‘italic’,fontsize = 12,col = “black”),
annoTerm.data = enrich,
line.side = “left”,
go.col = rep(ggsci::pal_d3()(8),each = 5),
go.size = “pval”,
mulGroup = c(2,2,2),
mline.col = c(ggsci::pal_lancet()(3)))
dev.off()
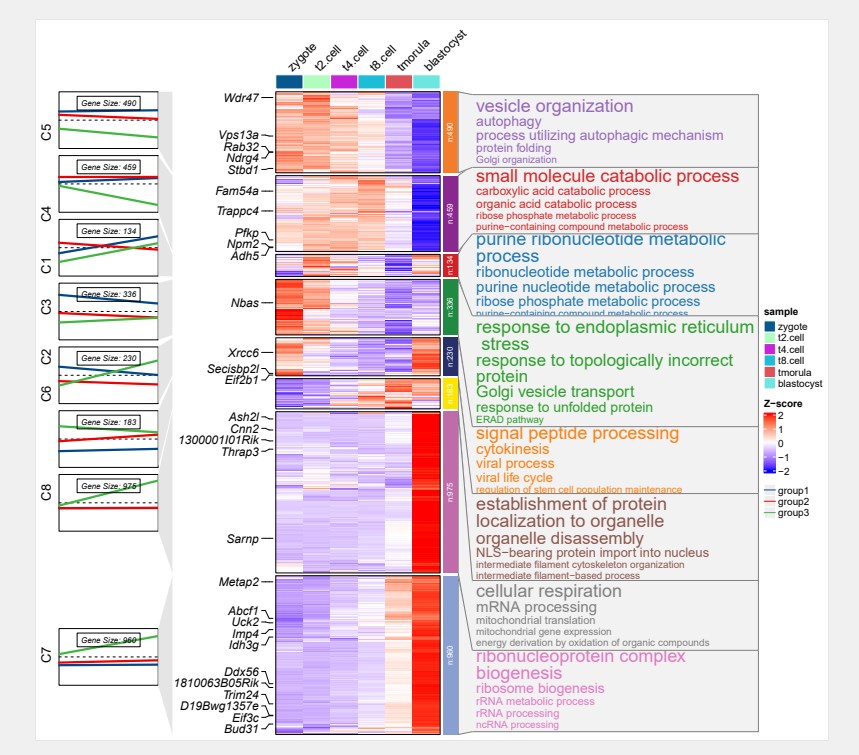
看起来绘图效果非常不错yyds,用起来非常方便,今天小果的分享就到这里,有需要的可以借鉴学习,下期再见。