重磅来袭,只需5分钟完全掌握单细胞多样本细胞互作分析
在上一期我们进行了单细胞单样本细胞互作分析,,我们为何要做单细胞互作分析?许多生物进程和细胞通讯息息相关,比如器官形成,胚胎发育,癌症发生发展,炎症反应,药物作用和耐药研究等;该分析在单细胞分析中具有很高的重要性,该分析属于单细胞分析中的高级分析内容,在文章中添加该分析会使你的文章增色不少,今天小果想尝试多样本细胞互作分析,还是利用CellChat 包进行分析,教程可以参考官网(https://htmlpreview.github.io/?https://github.com/sqjin/CellChat/blob/master/tutorial/Comparison_analysis_of_multiple_datasets.html),如果觉得推文不错,点赞加关注奥,话不多说开始今天的分享,代码如下:
- 安装需要的R包
library(devtools)
devtools::install_github(“sqjin/CellChat”)
install.packages(“magrittr”)
install.packages(“patchwork”)
install.packages(“tidyverse”)
BioCmanager::install(“Seurat”)
- 导入需要的R包
library(CellChat)
library(patchwork)
library(magtittr)
library(tidyverse)
library(Seurat)
- 读取示例数据
#该数据为前期保存的单细胞数据分析结果
immune.combined<- readRDS(“immune.combined.rds”)
- Chellchat多样本分析
## STIM组样本chellchat分析
stim.object <- subset(immune.combined,stim==”STIM”)
stim.data.input <- GetAssayData(stim.object, assay = “RNA”, slot = “data”)
stim.meta <- stim.object@meta.data[,c(“seurat_annotations”, “stim”)]
stim.meta$CellType %<>% as.vector(.)
#创建CellChat对象
stim.cellchat <- createCellChat(object = stim.data.input)
stim.cellchat <- addMeta(stim.cellchat, meta = stim.meta)
stim.cellchat <- setIdent(stim.cellchat, ident.use = “seurat_annotations”)
cellchat <- createCellChat(object = data.input, meta = meta, group.by = “labels”)
# CellChat提供的人的配受体数据库
stim.cellchat@DB <- CellChatDB.human
stim.cellchat <- subsetData(stim.cellchat)
#使用多线程进行计算
future::plan(“multisession”, workers = 10)
#用于细胞-细胞通信分析的表达数据预处理
stim.cellchat <- identifyOverExpressedGenes(stim.cellchat)
stim.cellchat <- identifyOverExpressedInteractions(stim.cellchat)
stim.cellchat <- projectData(stim.cellchat, PPI.human)
#在信号通路水平推断细胞间通信
stim.cellchat <- computeCommunProb(stim.cellchat)
#计算聚合的细胞间的通信网络
stim.cellchat <- aggregateNet(stim.cellchat)
#保存cellchat分析结果
saveRDS(stim.cellchat,”stim.cellchat.rds”)
## CTRL组样本Cellchat分析
ctrl.object <- subset(immune.combined,stim==”CTRL”)
ctrl.data.input <- GetAssayData(ctrl.object, assay = “RNA”, slot = “data”)
ctrl.meta = ctrl.object@meta.data[,c(“seurat_annotations”, “stim”)]
ctrl.meta$CellType %<>% as.vector(.)
ctrl.cellchat <- createCellChat(object = ctrl.data.input)
ctrl.cellchat <- addMeta(ctrl.cellchat, meta = ctrl.meta)
ctrl.cellchat <- setIdent(ctrl.cellchat, ident.use = “seurat_annotations”)
ctrl.cellchat@DB <- CellChatDB.human
ctrl.cellchat <- subsetData(ctrl.cellchat)
future::plan(“multisession”, workers = 10)
#用于细胞-细胞通信分析的表达数据预处理
ctrl.cellchat <- identifyOverExpressedGenes(ctrl.cellchat)
ctrl.cellchat <- identifyOverExpressedInteractions(ctrl.cellchat)
ctrl.cellchat <- projectData(ctrl.cellchat, PPI.human)
ctrl.cellchat <- computeCommunProb(ctrl.cellchat)
ctrl.cellchat <- computeCommunProbPathway(ctrl.cellchat)
#计算聚合的细胞间的通信网络
ctrl.cellchat <- aggregateNet(ctrl.cellchat)
#保存cellchat分析结果
saveRDS(ctrl.cellchat,”ctrl.cellchat.rds”)
- 加载两个数据集的Cellchat对象,然后合并在一起
object.list <- list(CTRL = ctrl.cellchat, STIM = stim.cellchat)
cellchat <- mergeCellChat(object.list, add.names = names(object.list))
- 进行互作次数和互作强度分析
gg1 <- compareInteractions(cellchat, show.legend = F, group = c(1,2))
gg2 <- compareInteractions(cellchat, show.legend = F, group = c(1,2), measure = “weight”)
pdf(“compareInteractions.pdf”,height = 5,width = 8)
gg1 + gg2
dev.off()
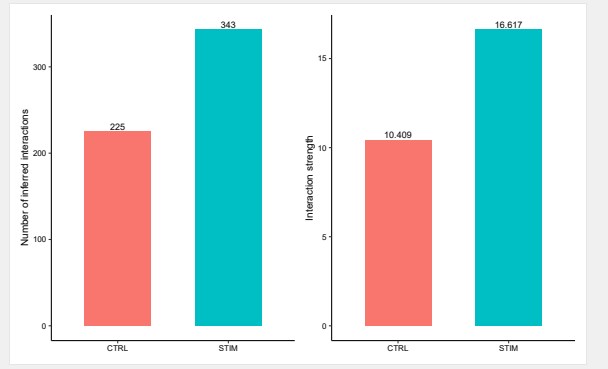
注:两个分组的互作次数和互作强度柱状图
- 不同细胞群相互作用次数和作用强度差异
#网络图绘制
pdf(“netVisual_diffInteraction.pdf”,height = 10,width = 10)
par(mfrow = c(1,2), xpd=TRUE)
netVisual_diffInteraction(cellchat, weight.scale = T)
netVisual_diffInteraction(cellchat, weight.scale = T, measure = “weight”)
dev.off()
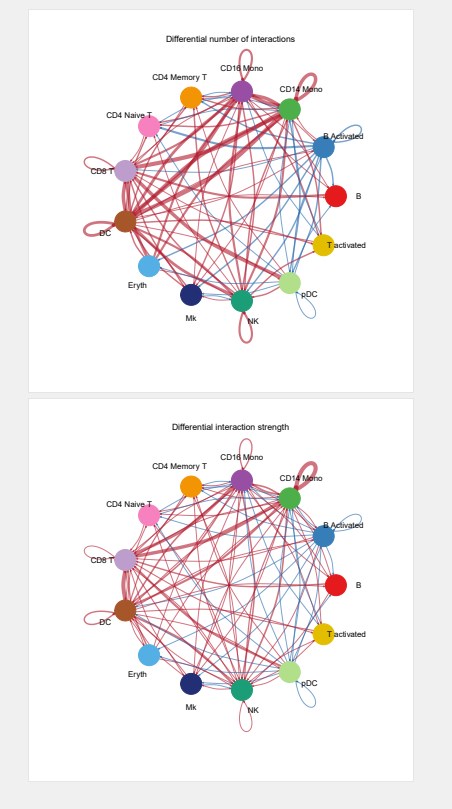
#热图绘制
gg1 <- netVisual_heatmap(cellchat)
gg2 <- netVisual_heatmap(cellchat, measure = “weight”)
pdf(“netVisual_heatmap.pdf”,height = 5,width = 8)
gg1 + gg2
dev.off()
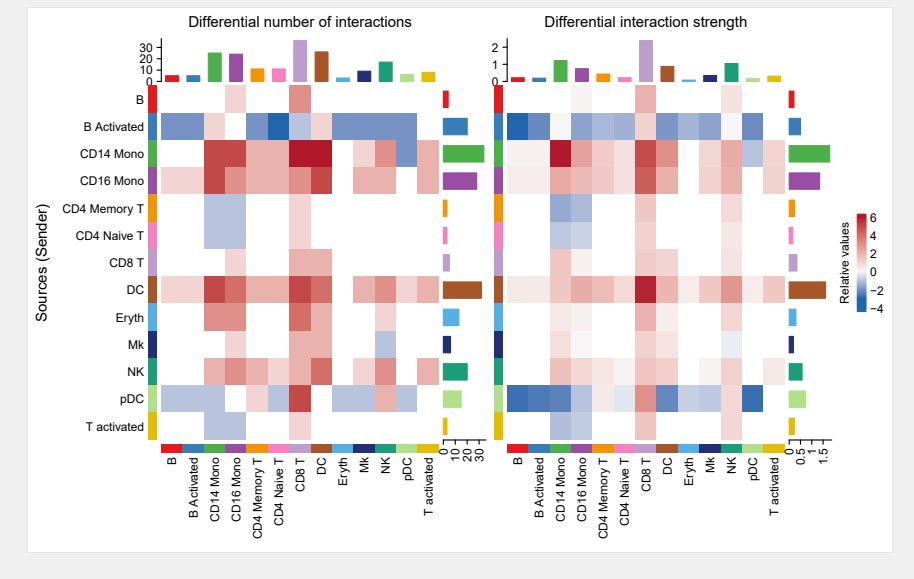
#不同数据集的互作网络图
pdf(“netVisual_diffInteraction.pdf”,height = 10,width = 10)
weight.max <- getMaxWeight(object.list, attribute = c(“idents”,”count”))
par(mfrow = c(1,2), xpd=TRUE)
for (i in 1:length(object.list)) {
netVisual_circle(object.list[[i]]@net$count, weight.scale = T, label.edge= F, edge.weight.max = weight.max[2], edge.width.max = 12, title.name = paste0(“Number of interactions – “, names(object.list)[i]))
}
dev.off()
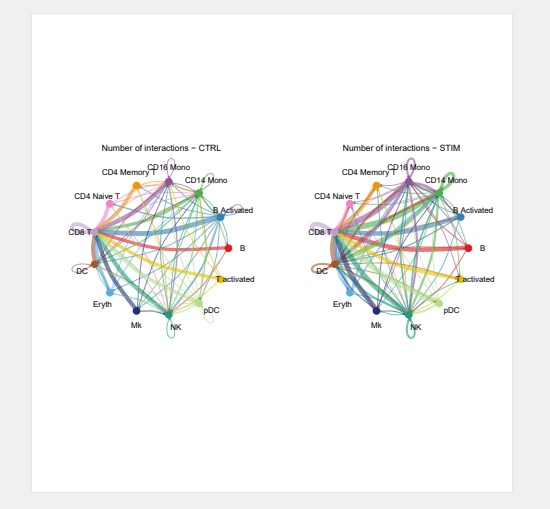
#识别上调和下调的受体-配体对
pdf(“netVisual_bubble.pdf”,height = 10,width = 10)
netVisual_bubble(cellchat, sources.use = 4, targets.use = c(5:11), comparison = c(1, 2), angle.x = 45)
dev.off()
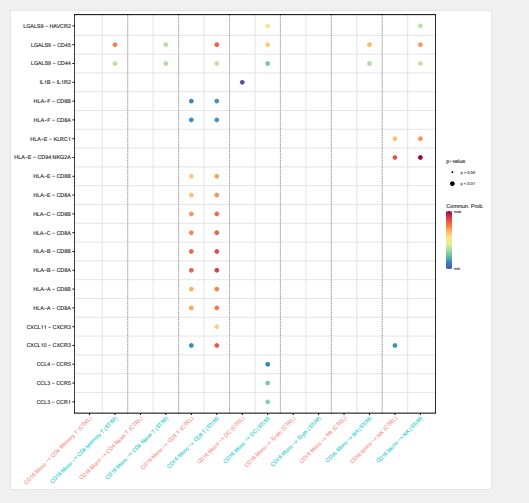
今天小果对单细胞多样本细胞互作分析的分享就到这里,干货满满奥,有需要的可以借鉴学习奥,进行该分析需要熟练掌握单细胞分析流程,单细胞相关分析内容可以尝试使用本公司新开发的云平台生物信息分析小工具,零代码完成分析,云平台网址:http://www.biocloudservice.com/home.html,包括单细胞分析(http://www.biocloudservice.com/366/366.php),单细胞数据绘制小提琴图(http://www.biocloudservice.com/788/788.php),绘制单细胞tSNE图(http://www.biocloudservice.com/229/229.php)等单细胞分析相关小工具,今天小果的分享就到这里,欢迎小伙伴和小果一起讨论学习奥。