重磅来袭,只需5分钟完全掌握单细胞多样本细胞互作分析

library(devtools)devtools::install_github("sqjin/CellChat")install.packages("magrittr")install.packages(“patchwork”)install.packages(“tidyverse”)BioCmanager::install(“Seurat”)
library(CellChat)library(patchwork)library(magtittr)library(tidyverse)library(Seurat)
#该数据为前期保存的单细胞数据分析结果immune.combined<- readRDS("immune.combined.rds")
## STIM组样本chellchat分析stim.object <- subset(immune.combined,stim=="STIM")stim.data.input <- GetAssayData(stim.object, assay = "RNA", slot = "data")stim.meta <- stim.object@meta.data[,c("seurat_annotations", "stim")]stim.meta$CellType %<>% as.vector(.)#创建CellChat对象stim.cellchat <- createCellChat(object = stim.data.input)stim.cellchat <- addMeta(stim.cellchat, meta = stim.meta)stim.cellchat <- setIdent(stim.cellchat, ident.use = "seurat_annotations")cellchat <- createCellChat(object = data.input, meta = meta, group.by = "labels")# CellChat提供的人的配受体数据库stim.cellchat@DB <- CellChatDB.humanstim.cellchat <- subsetData(stim.cellchat)#使用多线程进行计算future::plan("multisession", workers = 10)#用于细胞-细胞通信分析的表达数据预处理stim.cellchat <- identifyOverExpressedGenes(stim.cellchat)stim.cellchat <- identifyOverExpressedInteractions(stim.cellchat)stim.cellchat <- projectData(stim.cellchat, PPI.human)#在信号通路水平推断细胞间通信stim.cellchat <- computeCommunProb(stim.cellchat)#计算聚合的细胞间的通信网络stim.cellchat <- aggregateNet(stim.cellchat)#保存cellchat分析结果saveRDS(stim.cellchat,"stim.cellchat.rds")## CTRL组样本Cellchat分析ctrl.object <- subset(immune.combined,stim=="CTRL")ctrl.data.input <- GetAssayData(ctrl.object, assay = "RNA", slot = "data")ctrl.meta = ctrl.object@meta.data[,c("seurat_annotations", "stim")]ctrl.meta$CellType %<>% as.vector(.)ctrl.cellchat <- createCellChat(object = ctrl.data.input)ctrl.cellchat <- addMeta(ctrl.cellchat, meta = ctrl.meta)ctrl.cellchat <- setIdent(ctrl.cellchat, ident.use = "seurat_annotations")ctrl.cellchat@DB <- CellChatDB.humanctrl.cellchat <- subsetData(ctrl.cellchat)future::plan("multisession", workers = 10)#用于细胞-细胞通信分析的表达数据预处理ctrl.cellchat <- identifyOverExpressedGenes(ctrl.cellchat)ctrl.cellchat <- identifyOverExpressedInteractions(ctrl.cellchat)ctrl.cellchat <- projectData(ctrl.cellchat, PPI.human)ctrl.cellchat <- computeCommunProb(ctrl.cellchat)ctrl.cellchat <- computeCommunProbPathway(ctrl.cellchat)#计算聚合的细胞间的通信网络ctrl.cellchat <- aggregateNet(ctrl.cellchat)#保存cellchat分析结果saveRDS(ctrl.cellchat,"ctrl.cellchat.rds")
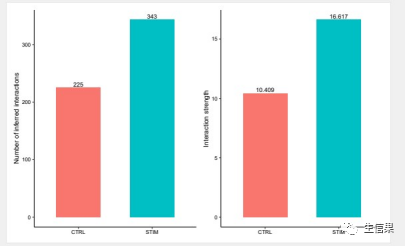
object.list <- list(CTRL = ctrl.cellchat, STIM = stim.cellchat)cellchat <- mergeCellChat(object.list, add.names = names(object.list))6. 进行互作次数和互作强度分析gg1 <- compareInteractions(cellchat, show.legend = F, group = c(1,2))gg2 <- compareInteractions(cellchat, show.legend = F, group = c(1,2), measure = "weight")pdf("compareInteractions.pdf",height = 5,width = 8)gg1 + gg2dev.off()
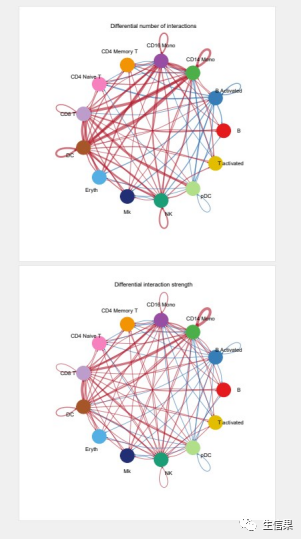
pdf("netVisual_diffInteraction.pdf",height = 10,width = 10)par(mfrow = c(1,2), xpd=TRUE)netVisual_diffInteraction(cellchat, weight.scale = T)netVisual_diffInteraction(cellchat, weight.scale = T, measure = "weight")dev.off()
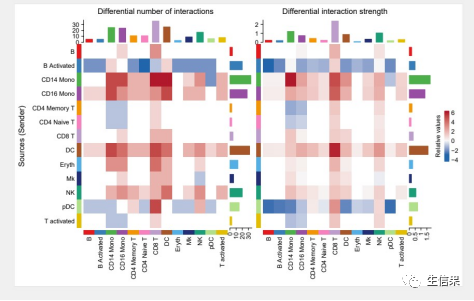
gg1 <- netVisual_heatmap(cellchat)gg2 <- netVisual_heatmap(cellchat, measure = "weight")pdf("netVisual_heatmap.pdf",height = 5,width = 8)gg1 + gg2dev.off()
pdf("netVisual_diffInteraction.pdf",height = 10,width = 10)weight.max <- getMaxWeight(object.list, attribute = c("idents","count"))par(mfrow = c(1,2), xpd=TRUE)for (i in 1:length(object.list)) {netVisual_circle(object.list[[i]]@net$count, weight.scale = T, label.edge= F, edge.weight.max = weight.max[2], edge.width.max = 12, title.name = paste0("Number of interactions - ", names(object.list)[i]))}dev.off()

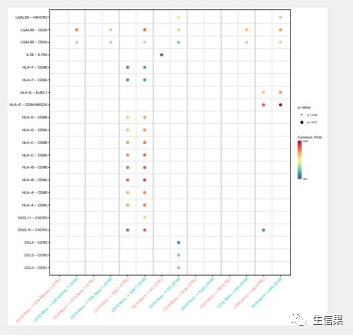
pdf("netVisual_bubble.pdf",height = 10,width = 10)netVisual_bubble(cellchat, sources.use = 4, targets.use = c(5:11), comparison = c(1, 2), angle.x = 45)dev.off()
生信人R语言学习必备
立刻拥有一个Rstudio账号
开启升级模式吧
(56线程,256G内存,个人存储1T)

